Once a vague scapegoat for a variety of ills, increasing evidence suggests a condition known as “leaky gut” – in which microbes and other molecules seep out of the intestines – may be more common, and more harmful, than previously thought.
Leaky gut is most often experienced by older people, patients with cancers or other chronic ailments, and people with especially stressful lifestyles. Stressors break down the zipper-like junctions between the cells that form the gut lining. Microbes and molecules that subsequently leak out through these cell gaps can trigger an immune response, contributing to a variety of diseases driven by chronic inflammation, such as inflammatory bowel disease, dementia, atherosclerosis, liver fibrosis, cancers, diabetes and arthritis.
Yet there isn’t a great way for clinicians to tell who has leaky gut and who does not – and there are no treatments to fix it.
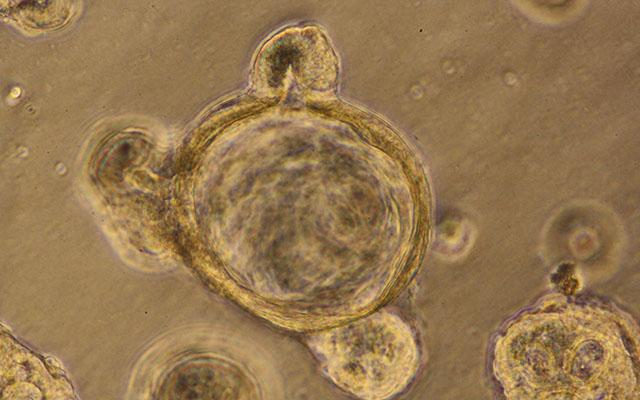
University of California San Diego School of Medicine researchers are now able to simulate leaky gut conditions for the first time, using 3D models of human intestines generated from patient cells. These small organoids, or “mini-guts,” have revealed new biomarkers that help define what a leaky gut looks like – molecular signals that could one day help clinicians better diagnose the condition, track its progression and evaluate the success or failure of treatments. The team also used the model to explore a potential pathway for tightening leaky guts with a common, available medication.
[ad_336]
The study, publishing February 10, 2020 in Life Science Alliance, was led by first author Pradipta Ghosh, MD, professor of cellular and molecular medicine at UC San Diego School of Medicine and Moores Cancer Center, and senior author Soumita Das, PhD, associate professor of pathology at UC San Diego School of Medicine.
Ghosh and colleagues had previously identified a specialized molecular mechanism that helps tighten gut lining junctions – a mechanism they dubbed the stress-polarity signaling pathway. They discovered that the junctions come undone when the pathway is under stress. They also found hints that the diabetes drug metformin might help activate the pathway, tightening up the junctions. But these fundamental discoveries were made in cell lines grown in petri dishes, without relevance in humans – at least not yet.
So Ghosh, Das and team turned to the next best thing to a human test subject: 3D gut organoids derived from patients’ intestines. The lining of the gut is not smooth, but more akin to rough terrain, with many peaks and valleys. At the bottom of each valley, known as a crypt, live small reservoirs of stem cells. To produce gut organoids, the researchers collected a tiny amount of these stem cells from patient biopsies and grew them in the lab. As they do in a person’s gut, the stem cells differentiated, or specialized, into the four different cell types that make up the gut lining. But in the lab, outside the body, they rolled up into a ball and formed crypts. In other words, “mini-guts.”
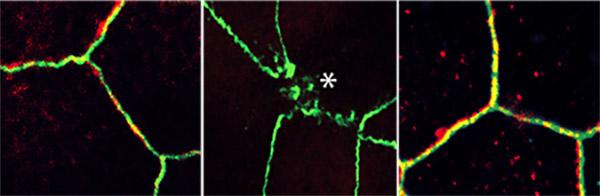
To simulate leaky gut conditions, the researchers rolled open the mini-gut balls to expose the surface of the intestinal lining. Then they sprinkled on several types of bacteria, which stressed the gut lining junctions, causing them to fall apart.
With this new model, Ghosh, Das and team confirmed that the junctions between cells are controlled by the previously identified stress-polarity signaling pathway. They discovered that the pathway keeping the gut lining intact begins to break down with aging and as colorectal tumors develop. They also measured an increase in markers of inflammation as the gut barrier began to fail.
[rand_post]
But this stress-polarity signaling pathway can be restored, the team found. The diabetes drug metformin activates AMPK, an enzyme that plays a key role in the stress-polarity signaling pathway. The researchers demonstrated that metformin strengthens the junctions of mini-guts, tightening the lining back up when stressed by bacteria, aging or tumor formation. One measure of stress-polarity signaling pathway strength is the levels of a molecule called occludin. In Ghosh and Das’ experiments, metformin increased occludin levels as much as six-fold compared to untreated cells.
Since each is derived from a different person, mini-guts vary in terms of their underlying genetics and epigenetics. That’s a strength, said Ghosh, but also a limitation.
“Lots of research is done in mice that are inbred so that they are genetically identical, all in the same cage, eating the same diet, in order to remove these variables from the studies,” she said. “But lab mice are far more standardized than the same human from day to day, or patients we see in the clinics. Here, our model is a better representation of humanity. On the other hand, it also means that each organoid is its own unique experiment. We have to test many organoids to be able to make any claim, which we did in our study.”
Researchers next want to take a closer look at the diseases driven by leaky gut. They also plan to test various ways to tighten junctions in the context of aging, inflammatory bowel disease, cancer and other conditions to see if they can reduce or prevent initiation and progression of these diseases.
“I think you’d be hard pressed to find a disease in which systemic inflammation is not a driver,” Das said. “That’s why, even though there are so many things we still don’t know, we’re excited about the broad potential this model and these findings open for developing personalized leaky gut therapeutics that target AMPK and the stress-polarity signaling pathway.”