Neuroscientists and machine learners from University of Zurich and ETH Zurich have introduced a novel artificial intelligence (AI)-based method to develop a brain atlas through deep learning.
The researchers published their results on June 10, 2019 In Nature Machine Intelligence.
Brain atlases for mice, humans and other species are not available for every developmental stages which brings a great challenge for the neuroscience community in mapping their brain regions of interest. Another challenge is to go through a time consuming step; that is, to select the brain section image and manually register it against the corresponding reference atlas image. This approach can easily takes weeks even months depending on the size of brain imaging dataset.
Authors propose that the task of registering images of brain sections against a standard reference atlas can also be achieved by using a deep neural network (DNN) to segment regions in the brain through feature engineering (figure 1). Therefore, they propose a different approach to the classical problem of brain image registration, which relies on segmentation. Instead of relying on a reference atlas to be chosen and mapped by human experts, a trained deep neural network is able to segment the brain regions (SeBRe) with an accuracy close to human experts.
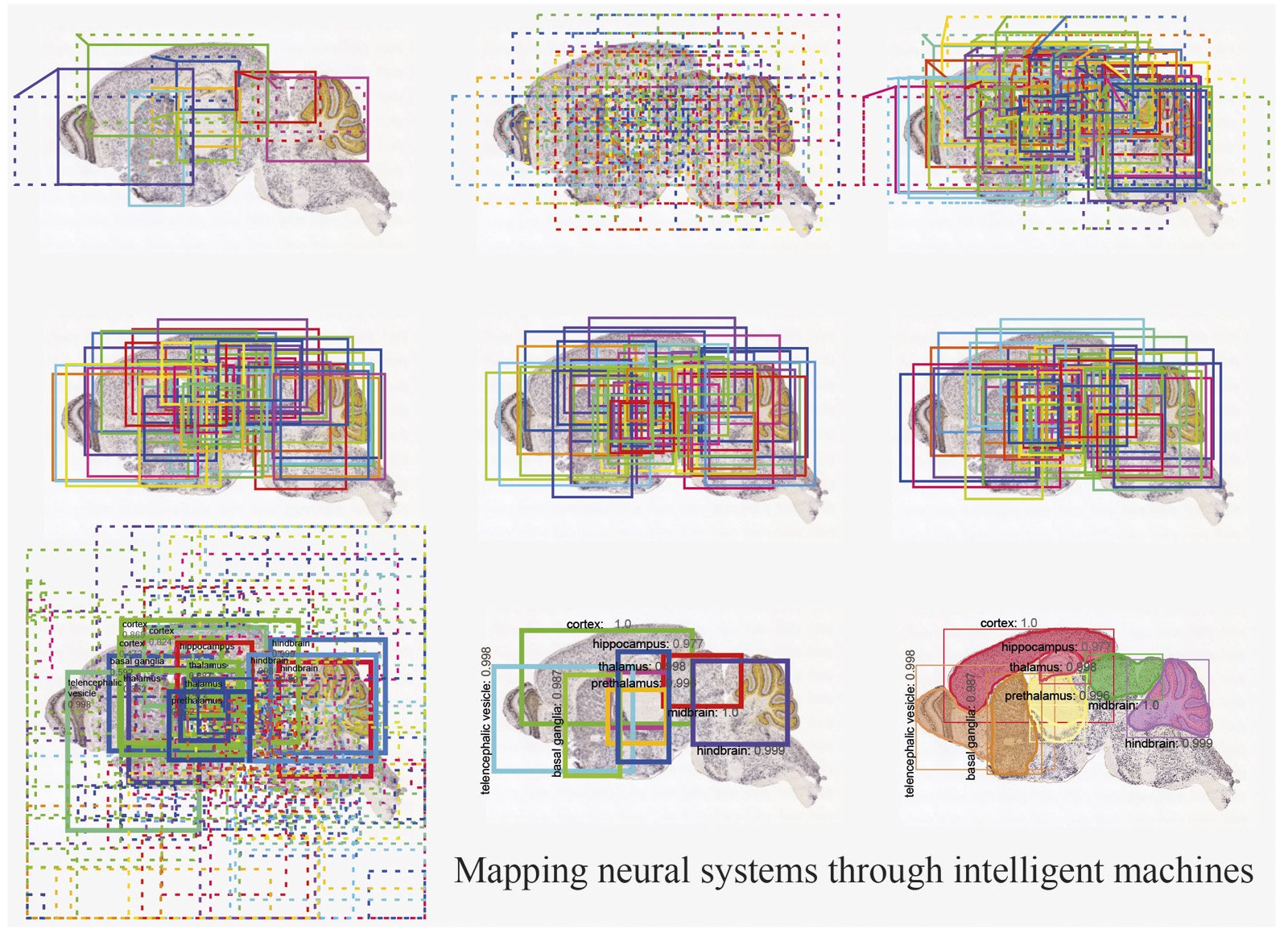
The authors demonstrate the performance of SeBRe on mouse brain images labelled with a variety of genetic markers as well as human magnetic resonance imaging (MRI) images. SeBRe takes the brain sections and their human-annotated binary (ground-truth) masks as input and after training it segments the brain regions in an unseen brain section image, without relying on the reference atlas.
[ad_336]
SeBRe is scale invariant; that is, it performs equally well in segmenting brain images that vary substantially in the relative sizes of the brain regions. This is crucial not only for accurately segmenting regions at different stages of development, but also in tissue sections in which the proper geometry of the brain has been compromised due to any methodological issues (for example, during brain tissue slicing) or even pathological deformities (such as neurodegeneration in Alzheimer’s disease). This is demonstrated by the fact that although SeBRe is trained only on a mice Post-natal (P) day 14 brain image dataset, the network performs equally well on other developmental ages, including P4, P28 and P56. Furthermore, the performance of SeBRe is equally well on brain sections obtained through a different imaging modality (figure 2), which points towards the generalizability of SeBRe in segmenting brain regions in a variety of bioimaging modalities.
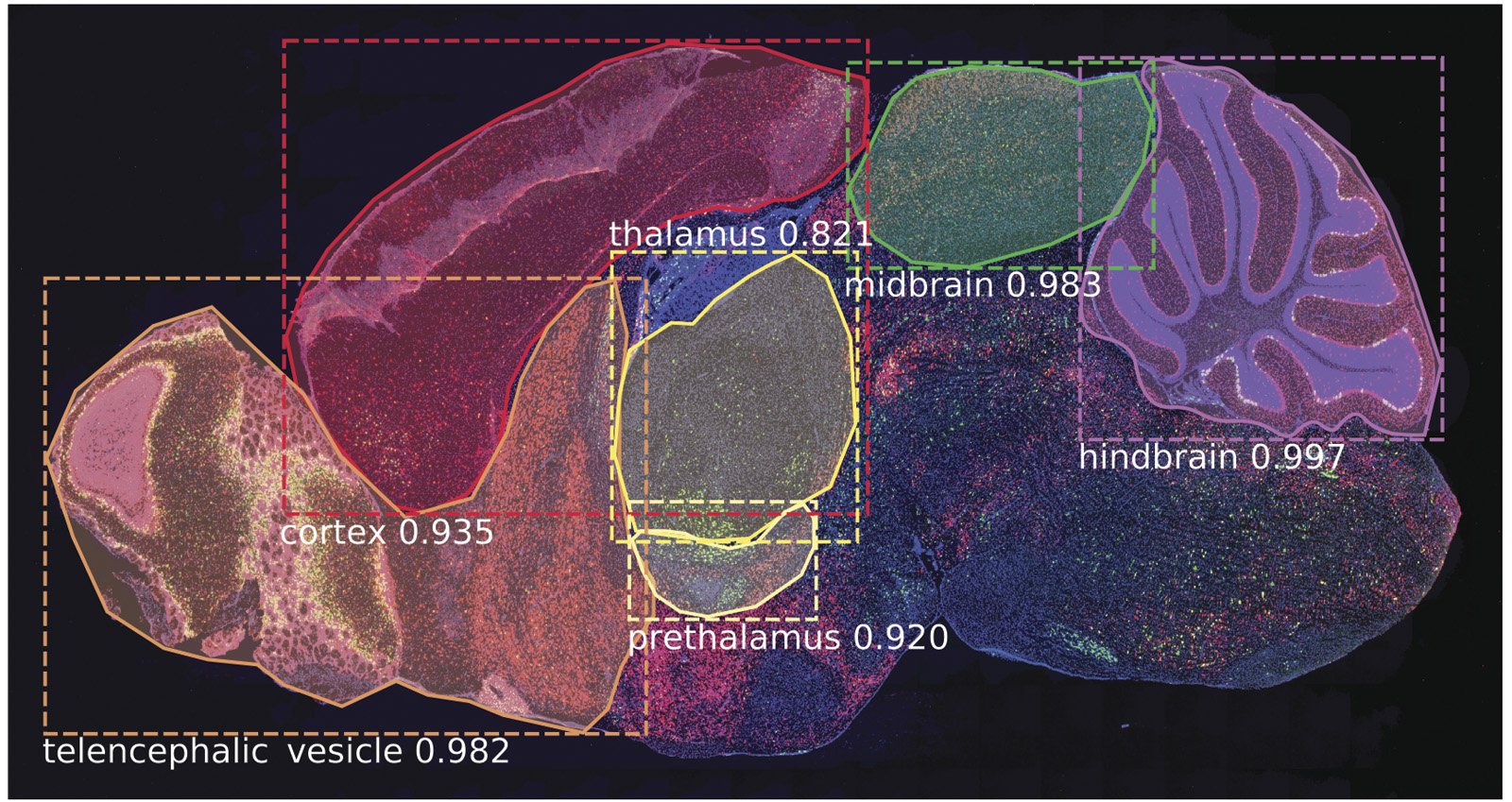
[rand_post]
“We demonstrate the performance of SeBRe in segmenting eight regions in mouse and human brains. Our goal is to a utilize the power of AI and develop a high-throughput system to generate the precise maps of brain regions and sub-regions which the neuroscientists can easily navigate through, much like we virtually navigate in different places and streets through Google Maps.” Iqbal said.