An international team consisting of researchers from the Macromolecular Complexes in DNA Damage Response Group, headed by Óscar Llorca at the Spanish National Cancer Research Centre (CNIO), and the group led by Sebastian Geibel at the University of Würzburg (Germany), worked together in a multidisciplinary effort to obtain an accurate 3D model of the mechanism used by the bacterium Mycobacterium tuberculosis to block the immune response when infecting an organism, so the infection can success.
This long-awaited finding among members of the tuberculosis research community, which may help revolutionize the search for new treatments against this and other bacterial diseases, is published in the prestigious journal Nature. In a time when more and more bacteria are developing resistance to antibiotics that are used to fight them – one of the great challenges to face according to the World Health Organization (WHO) -, and considering that this disease is still a great threat in underdeveloped countries with inadequate health care systems, ending the tuberculosis epidemic is one of the most urgent health issues of the Sustainable Development Goals established by the United Nations Organization (UN) for the year 2030.
Tuberculosis is an infectious disease that has a high mortality rate: it is one of the top ten causes of death worldwide and especially affects people with HIV and other pathologies that affect the immune system. According to data from WHO, 10 million people contracted tuberculosis in 2017 worldwide; 1.6 million of them died. Since the current treatment has been in use for 40 years and new antibiotic-resistant strains of the disease are emerging, the need for new therapeutic strategies is urgent. To do so, it is necessary to have an in-depth understanding of the mechanisms that the bacteria use to cause the disease.
Bacteria against the immune system
When an organism is infected by M. tuberculosis, the immune system launches a complex response to destroy it. The bacterium has evolved several sophisticated mechanisms to undermine the immune system and help to escape their fate. The bacterium uses a secretion system – a protein complex located in its membrane – to inject certain virulence factors into the cells of the immune system. These factors are molecules with the task to paralyze the defensive response of the immune cells, so that bacteria, instead of being destroyed, are given free rein to continue infecting the body.
[ad_336]
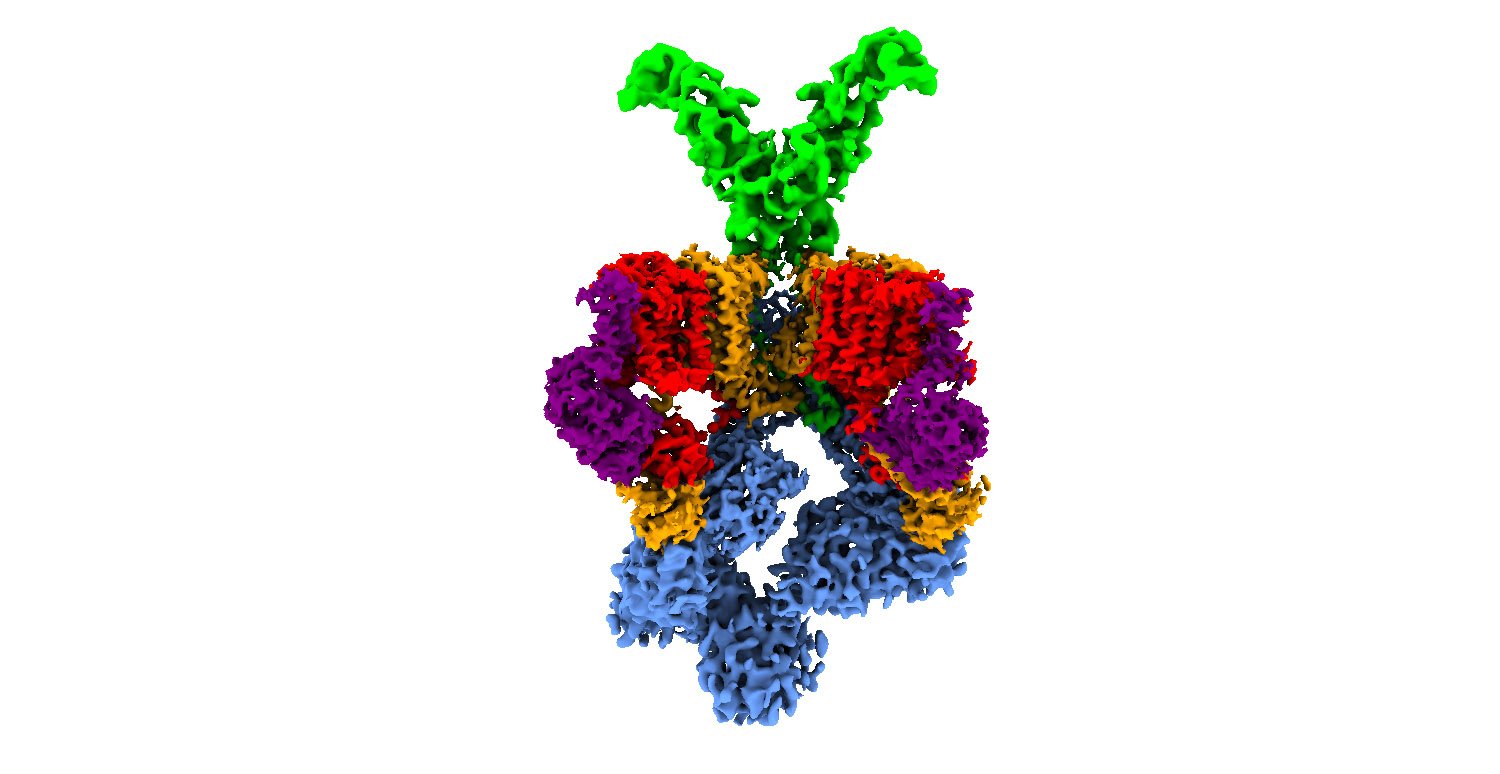
The structure and operation mechanism of the M. tuberculosis secretion system, called T7SS (type VII secretion system), had not been known in detail. Until now, only very low-resolution structural information had been obtained, which showed a structure in the form of a hexamer (six-pointed star) whose center serves as a channel through which the bacterium ejects the virulence factors. The lack of information on T7SS and on how it works at the atomic level has prevented progress in achieving new therapeutic strategies against tuberculosis based on attacking the secretion system.
Now, researchers Óscar Llorca and Ángel Rivera-Calzada of CNIO, who contributed their expertise in cryo-electron microscopy (cryo-EM) and digital image processing, and Sebastian Geibel and Nikolaos Famelis from the University of Würzburg, experts in bacterial secretion systems, combined forces to unravel this puzzle and they managed to describe in detail the T7SS at the atomic level. The researchers worked with a very similar bacterium, M. smegmatis, which is used in research as a model to study M. tuberculosis and that shares the same secretion system with it. The work has shown that T7SS is a sophisticated nanomachine, in which several proteins cooperate to inject the virulence factors produced by the bacterium into the cells of the immune system.

Towards a new generation of drugs
Recent developments have transformed cryo-EM into an extremely powerful technology that allows high-resolution imaging of molecular structures. This technique allows accelerating the obtaining of structural information that with other techniques would require large volumes of the sample or its crystallization. With this technique, molecular biology and biomedicine are taking a huge qualitative leap that is expected to revolutionize the development of treatments of diseases.
[rand_post]
In this paper, and thanks to the multiple images of the T7SS obtained and processed using cryo-EM researchers of CNIO and the University of Würzburg have at long last pinpointed all the components of T7SS, have elucidated the function of some of them that remained unknown, have modeled its three-dimensional structure and, based on all this information, have proposed an operating mechanism.
“We were able to see that the components that until now appeared blurred with other techniques are in fact elements that are in constant motion,” explains Llorca. “Thus, we saw that the hexamer of T7SS is composed of a subcomplex of 4 proteins and that 6 identical copies of this subcomplex are needed to shape the six-pointed star around a central pore, through which the virulence factors that block the defensive response of the infected organism are ejected.” Subsequently, the proposed mechanism was successfully tested by the Würzburg University group using different mutated versions of the system.
The system used by the German group to test the mechanism is also of great interest to the research community: “It will be very useful to test the effect of new molecules directed against this secretion mechanism, which the bacteria of the genus Mycobacterium need to successfully carry out the infection,” explains Rivera-Calzada.
This multidisciplinary study opens a new field to explore for diseases caused by bacterial infections, since knowing the three-dimensional structure of the different bacterial secretion systems will allow exploring new compounds that block secretion. In point of fact, in the next steps of this research, the team of CNIO and the University of Würzburg will try to study in greater depth how the secretion process itself occurs in Mycobacterium, to open the way to the design of molecules that can block it.