To understand how individual molecules play their roles in biological processes inside the cells they are synthesized in, researchers have developed super-resolution microscopy methods to visualize them at the single-molecule level. However, to investigate their functions, ultimately, they would also like to be able to modify them individually at this high resolution. While the visualization of single molecules has made great strides in recent years, thus far it has remained challenging to directly modify them in a controlled, molecule-to-molecule fashion.
[ad_336]
Now, reported in Nature Chemistry, researchers at Harvard’s Wyss Institute for Biologically Inspired Engineering and Harvard Medical School (HMS), have developed “Action-PAINT”, a method that combines the team’s real-time DNA-PAINT super-resolution microscopy approach with a single-molecule labeling strategy at a desired location within synthetic nanostructures or intact cells. This approach could be further developed to allow researchers to stimulate or inhibit the functions of individual molecules and study the consequences for normal biological and disease-related processes in real time and super-resolution.
“Super-resolution imaging methods have allowed us to ‘see the previously invisible.’ By coupling our DNA-PAINT super-resolution microscopy method with a cross-linking approach, we now can also ‘touch the previously inaccessible’ by attaching a physical handle to individually observable molecules right at the time of visualization,” said Wyss Institute Core Faculty member Peng Yin, Ph.D., who led the study. Yin is also a co-leader of the Institute’s Molecular Robotics Initiative, and Professor of Systems Biology at HMS.
Yin’s team developed a two-step approach that first visualizes single proteins or other molecules at super-resolution, and then transfers a molecular label to the desired target site. Optionally, the researchers can confirm successful transfers with an additional round of super-resolution imaging.
[rand_post]
The first step of single-molecule imaging relies on the DNA-PAINT method that enabled the team to first determine the precise localization of molecules or molecular features that are spatially separated by distances well below the diffraction limit of light, and therefore invisible to most microscopes. The researchers first attached a short “docking strand” of DNA to the target that serves as a binding site for a complementary “imager strand” carrying a fluorescent dye. Because the imager strand binds with a programmable on-off rate, it produces defined “blinking” events which can be observed using standard microscopes. To attach a physical label to the target, the researchers carried out their first super-resolution imaging step with a slightly more complex imager strand that also included a photo-inducible cross linker, which is able to chemically link docking and imager strand to each other when exposed to UV radiation, and an additional reporter sequence.
“A critical component of our new method is the precise control of the crosslinking laser, which works in synchrony with the super-resolution blinking sequence. This really converts our DNA-PAINT super-resolution microscopy from a passive imaging method to an active one, enabling real-time interaction between the researcher and individual molecular targets,” said co-first and co-corresponding author Mingjie Dai.
To enable this new capability, the team developed a software package that allowed them to first map out the precise locations of all molecular targets in an area of interest using the DNA-PAINT method, and then tightly synchronize the flashing of a UV with subsequent blinking events. “This way we could chemically bind the imager strand to custom-selected molecular targets, one by one, with molecular resolution – like a painter lays down his color, patch by patch in the pointillism style,” said Dai, Ph.D., a former postdoctoral fellow on Yin’s team, who currently is a Departmental Fellow at HMS’s Department of Systems Biology and Technology Development Fellow at the Wyss Institute.
[ad_336]
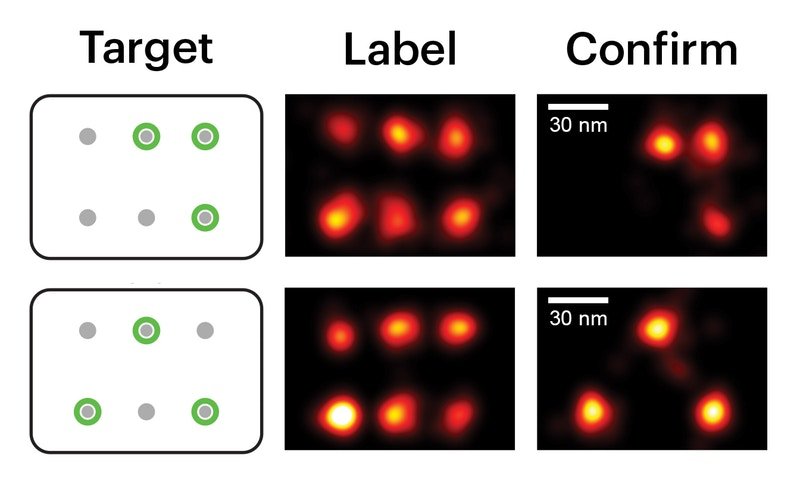
To demonstrate the effectiveness and selectivity of their approach, the team first used synthetic DNA nanostructures that exposed docking sites for imager strands in defined patterns.
“Putting Action-PAINT to work, we started by demonstrating its effectiveness labelling single molecule targets distanced merely 30 to 70nm from their identical neighbors with high on-target efficiencies,” said co-first author Ninning Liu, Ph.D., “and then further validated our method in fixed cells, where we selected and labeled microtubule proteins along cytoskeletal filaments, with different custom-defined patterns.” Liu is a postdoctoral fellow on Yin’s team.
The authors envision that Action-PAINT could be further developed into a broadly applicable tool that, for example, could help directly modify the activities of single membrane receptors on the surface of cells from where they direct cell behavior, or of ion channels that control the function of neuronal cells. In addition, the methods could allow the transfer of molecular handles to single proteins that may allow their extraction and purification together with other proteins they bind to naturally.
“Action-PAINT adds yet another level of functionality to the capabilities developed by Peng Yin’s team that can enable both molecular position-based functional protein interaction mapping inside individual cells, and biochemical analysis of these interactions once these molecules are isolated, which in the future could help to identify and/or validate new drug targets,” said Wyss Institute Founding Director Donald Ingber, M.D., Ph.D., who is also the Judah Folkman Professor of Vascular Biology at HMS, the Vascular Biology Program at Boston Children’s Hospital, and Professor of Bioengineering at Harvard’s John A. Paulson School of Engineering and Applied Sciences.
[rand_post]
The study was also authored by Sinem Saka, Ph.D., a postdoctoral fellow in Yin’s group at the Wyss Institute. It has been funded by the Molecular Robotics Initiative at Harvard’s Wyss Institute for Biologically Inspired Engineering, the National Institutes of Health, Office of Naval Research, and the National Science Foundation.